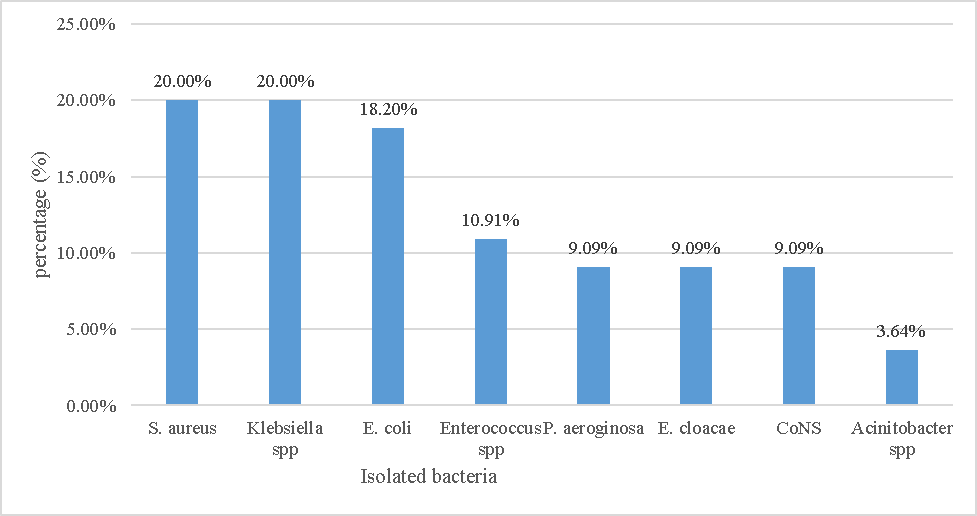
Figure 1 Frequency of bacterial species involved in surgical site infections in UoGCSH, Northwest Ethiopia, February - April 2020.
Bacterial profile and antimicrobial susceptibility patterns of isolates from postoperative surgical site infections and hospital environment samples
1Department of Medical Microbiology, University of Gondar, College of Medicine and Health Sciences, School of Biomedical and Laboratory Sciences, Gondar, Ethiopia.
2 Department of Pathology, College of Medicine and Health Science, Bahir Dar University, Ethiopia
3Department of Medical Microbiology, Amhara National Regional State Public Health Institute, Bahir Dar, Ethiopia
*Corresponding Author: Gizeaddis Belay: Email: gizebetaddis@gmail.com, Mobile Number +251 935 48 1335,
Background: The occurrence of microorganisms especially antimicrobial-resistant bacteria in health facilities can cause infections among admitted patients. This increases the treatment costs, prolonged hospital stays, and significant morbidity and mortality for postoperative patients. Currently, there is insufficient evidence of surgical site infection and multidrug-resistant bacteria. Therefore, continuous surveillance is necessary to guide an appropriate therapy for surgical site infection and the rational use of antimicrobial agents. Thus, this study provides updated information on information about the bacteria, Multi-Drug Resistance bacteria species responsible for postoperative surgical site infection, and the etiologic agents in hospital environments.
Objectives: This study assesses bacterial profile and antimicrobial susceptibility patterns in samples collected from postoperative surgical site infections and the hospital environment at the University of Gondar Comprehensive Specialized Hospital, Gondar; Northwest Ethiopia.
Materials and methods: A cross-sectional study was conducted among patients with postoperative surgical site infections and hospital environment samples from February 1 to April 30, 2020. All postoperative patients suspected of having surgical site infections and hospital environments were included in the study. A total of 202 samples (52 from wounds and 150 from the environment) were examined. Socio-demographic characteristics were collected using a structured questionnaire. Swab samples were obtained and inoculated onto MacConkey agar, Mannitol salt agar, Blood agar plates, and Chocolate agar by rolling the swab over the agar surfaces. The inoculated plates were then incubated at 37 °C for 24 to 48 hours. Air culture samples from Blood agar plates were also incubated at 37 °C for 24 hours. Antimicrobial susceptibility testing was conducted using the disk diffusion method on Muller Hinton agar. Data were entered and analyzed using Statistical Package for the Social Sciences version 20. Descriptive statistics were employed to present the findings through words, percentages, and tables.
Results: Of 52 wound samples from surgical site infection, the most frequent isolates were S. aureus and Klebsiella species, each accounting for 11 cases (20%), followed by E. coli with 10 cases (18.2%). Among the S. aureus isolates, 63.6% were methicillin-resistant. The overall rate of multidrug resistance was 31 cases (56.4%). Regarding hospital environmental samples, of 150 samples, the most commonly identified isolates were coagulase-negative S. aureus with 57 cases (47.5%), followed by S. aureus with 35 cases (29.2%). The overall rate of multidrug resistance was 66 cases (55.0%).
Conclusion: Staphylococcus aureus, Klebsiella species, and E. coli were identified as the most prevalent bacteria from postoperative surgical site infections, with hospital environments serving as potential reservoirs for these pathogens in the study area. High prevalence rates of methicillin-resistant and multidrug-resistant were observed among both clinical and hospital isolates in this study. However, Amikacin and Clindamycin demonstrated the highest effectiveness in inhibiting the in vitro growth of Gram-negative and Gram-positive bacterial isolates, respectively. Therefore, updating treatment guidelines based on hospital formularies and susceptibility patterns is crucial to prevent the further emergence and spread of multidrug-resistant bacterial pathogens. Additionally, infection prevention practices should be strengthened.
Keywords: antimicrobial susceptibility; Environmental sample; surgical site infection; Gondar; Ethiopia.
የጥናቱ ዳራ፡- በጤና ተቋማት ውስጥ፣ ረቂቅ ተሕዋስያን በተለይም ፀረ ተሕዋስያንን የሚቋቋሙ ባክቴሪያዎች መከሰት ወደ ተቋሙ በሚገቡ ታካሚዎች ላይ ኢንፌክሽን ሊፈጥር ይችላል። ይህ የሕክምና ወጪን፣ የሆስፒታል ቆይታዎችን እና ከቀዶ ጥገና በኋላ ለታካሚዎች ከፍተኛ የሆነ በበሽታ የመያዝና ሞት ይጨምራል። በአሁኑ ጊዜ፣ በቀዶ ጥገና ቦታ ስለሚከሰት ኢንፌክሽንና ብዙ መድሃኒቶችን የተላመዱ ባክቴሪያዎችን በተመለከተ ምንም ማስረጃ የለም። በቀዶ ጥገና ቦታ ለሚከሰት ኢንፌክሽን ተገቢውን ህክምና ለመስጠትና ምክንያታዊ የጸረተሕዋሲያን አጠቃቀምን ለመምራት የማያቋርጥ ክትትል አስፈላጊ ነው። ስለዚህ ይህ ጥናት ስለባክቴሪያ ማለትም ድኅረ ቀዶ ጥገና ቦታ ላይ ለሚከሰት ኢንፌክሽን ምክንያት የሆኑና ብዙ መድሃኒቶችን የተላመዱ ባክቴሪያዎች እና በሆስፒታል አካባቢ በሽታ አስተላላፊ ቁሶችን በተመለከተ ወቅታዊ መረጃ ይሰጣል።
የጥናቱ ዓላማዎች፡- ይህ ጥናት በሰሜን ምዕራብ ኢትዮጵያ፣ ጎንደር በሚገኘው የጎንደር ዩኒቨርሲቲ አጠቃላይ ስፔሻላይዝድ ሆስፒታል ውስጥ በቀዶ ጥገና ቦታ ላይ ከሚከሰት ኢንፌክሽን እና የሆስፒታል አካባቢ በተወሰዱ ናሙናዎች አማኝነት የባክቴሪያ ፕሮፋይል እና ፀረ ተህዋሲያን ተጋላጭነት ሁኔታን መገምገምን ያለመ ነው።
የጥናቱ ዘዴ፡- እ.አ.አ. ከየካቲት 1 አስከ ሚያዚያ 30፣ 2020 ድረስ በተወሰደ ናሙና በድኅረ ቀዶ ጥገና ቦታ በሚከሰት ኢንፌክሽንና በሆስፒታለ አካባቢ በታመሙ በሽተኞች ላይ ተሻጋሪ ጥናት ተካሂዷል። በቀዶ ጥገና ቦታ ለሚከሰት ኢንፌክሽን ተጋላጭ የሆኑ የድኅረ ቀዶጥገና ታማሚዎች ሁሉም በጥናቱ ተካትተዋል። በዚህ ጥናት 52 ቁስሎችና 150 አካባቢዎች በድምሩ 202 ናሙናዎች ተፈትሸዋል። የማኅበረ ሥነ ሕዝባዊ መገለጫዎች በዝግ የጽሑፍ መጠይቆች ተሰብስበዋል። ስዋብ (Swab) ናሙናዎች ተሰብስበው በማክኮንኪ አጋር፣ በማኒቶል ጨው አጋር፣ በደም አጋር ሳህን እና በቸኮሌት አጋር ውስጥ ስዋቡን በአጋር ላይ በማንከባለል ኢኖክሌት ተደርገዋል። ኢኖክሌት የተደረጉትን የአጋር ሳህኖችን በ37 ዲግሪ ሴንቲ ግሬድ ውስጥ ከ24-48 ሰዓታት በማቆየት ተላላፊ በሽታዎች እንዲያድጉ ተደርገዋል። የአየር ባህል የደም አጋር ሰሃንም በ 37 ዲግሪ ሴንቲ ግሬድ ውስጥ ለ 24 ሰዓታት እንዲቆዩ በማድረግ ተላላፊ በሽታዎች እንዲያድጉ ተደርገዋል። በሙለር ሂንተን አጋር ላይ ያለውን የዲስክ ስርጭት ቴክኒክ በመጠቀም የፀረ-ተህዋሲያን ተጋላጭነት ሙከራዎች ተካሂደዋል። መረጃው የገባ እና የተተነተነው በስታቲስቲክስ ፓኬጅ ለሶሻል ሳይንሶች ስሪት 20 በመጠቀም ነው። ግኝቶችን በቃላት፣ በመቶኛ እና በሰንጠረዦች ለማቅረብ ገላጭ ስታቲስቲክስ ጥቅም ላይ ውሏል።
የጥናቱ ውጤት፡- በቀዶ ሕክምና ቦታ ከሚከሰት ኢንፌክሽን የተገኙ በጣም የተለመዱ ተለይዎች (isolates) ኤስ. አውረስ (S. Aureus) እና ክለብሲየላ (Klebsiella) ዝርያዎች እያንዳንዳቸው 11(20 በመቶ) ፣ እና E.coli 10(18.2በመቶ) ነበሩ። ከኤስ. አውረስ (S. Aureus) ተለይዎች ውስጥ፣ 63.6 በመቶ ሜቲሲሊንን የተላመዱ ነበሩ። አጠቃላይ የብዝሃ-መድሀኒት መላመድ 31 (56.4በመቶ) ነበር። የሆስፒታል አካባቢያዊ ናሙናዎችን በተመለከተ፣ በጣም የተለመዱት ተለይዎች ኮአጉለስ የሌላቸው ኤስ. አውረስ (coagulase-negative S. Aureus) 57 (47.5በመቶ) እና S. Aureus 35 (29.2በመቶ) ናቸው። አጠቃላይ የብዝሃ-መድኀኒት መላመድ 66(55.0በመቶ) ነበር።
የጥናቱ ማጠቃለያና:- ይህ ጥናት ስቴፊሎኮከስ ኦውሬስ (Staphylococcus aureus) ፣ ክሌብሲየላ (Klebsiella) ዝርያዎች እና ኢ. ኮላይ (E. coli) ባክቴሪያዎች በድኅረ ቀዶ ጥገና ቦታ ለሚከሰቱ ኢንፌክሽኖች ዋና ዋና መንስኤዎች መሆናቸውንና የሆስፒታሉ አከባቢዎች በጥናቱ አካባቢ ላሉ በሽታ አምጪ ተሕዋሲያን እምቅ ማጠራቀሚያ ሆነው እንደሚያገለግሉ አሳይቷል። በተጨማሪም በዚህ ጥናት ውስጥ ከፍተኛ ሜቲሲሊንን የተላመደና ብዙ መድሃኒቶችን የመላመድ ስርጭት በሁለቱም በክሊኒካዊ እና በሆስፒታል ውስጥ ተስተውሏል። ይሁን እንጂ አሚካሲንና ክሊንዳሚሲን የግራም-አሉታዊ እና አወንታዊ የባክቴሪያ መነጠልን በብልቃጥ እድገትን ለመግታት በጣም ውጤታማ መድሃኒቶች ሆነው ተገኝተዋል። ስለሆነም የመድኀኒት የተላመደ በሽታ አምጪ ተህዋሲያን የበለጠ እንዳይከሰቱና እና እንዳይስፋፉ ለመከላከል በሆስፒታሉ ፎርሙላሪ እና በተጋላጭነት ሁኔታ ላይ በመመርኮዝ የፀረ-ተባይ መድኀኒቶች አጠቃቀም የሕክምና መመሪያዎች መዘመን አለባቸው። በተጨማሪም የኢንፌክሽን መከላከያ ዘዴዎችን ማጠናከር ያስፈልጋል።
ቁልፍ ቃላት፡- ፀረ-ተሕዋስያን ተጋላጭነት፣ የአካባቢ ናሙና፣ በቀዶ ጥገና ቦታ ይሚፈጠር ኢንፌክሽን
Healthcare-associated infections (HAIs), also known as "nosocomial" or "hospital-acquired" infections, are infections acquired within 48 hours of hospital admission or up to three days after discharge from the hospital or surgical center 1,2. Some examples of common healthcare-associated infections are catheter-associated urinary tract infections, ventilator-associated pneumonia, surgical site infections (SSI), and central line-associated bloodstream infections 3.
A surgical site infection is defined as an infection that occurs within 30 days of an operation at or near the surgical incision site, or within 1 year if an implant was placed. These infections can be classified into incisional SSI (superficial and deep) and organ/space SSI 4. It is one of the most common healthcare-associated infections 5,6. Approximately 80% to 90% of postoperative infections occur within 30 days following the operative procedure 7.
The preponderant bacteria most frequently associated with SSIs are Staphylococcus aureus, coagulase-negative Staphylococci (CoNS), Enterococcus species, Escherichia coli, Pseudomonas aeruginosa, Enterobacter species and Klebsiella pneumonia 8. These pathogens causing SSIs are believed to originate from the patient's own body (endogenous flora), contact with healthcare staff (cross-contamination), contaminated hospital environments, and surgical instruments (exogenous flora) 9,10. Contamination of the hospital environment contributes to the multiplication, dissemination, and transmission of pathogens to patients undergoing operative procedures 11. Transmission of these microorganisms to patients mainly occurs through contact with contaminated hospital surfaces, particularly through hand contact 12.
Multidrug-resistant bacteria such as Methicillin Resistant S. aureus (MRSA), Vancomycin-resistant Enterococci, and multidrug-resistant Gram-negative bacteria are common causes of postoperative SSI 7. Gram-positive bacteria like S. aureus can survive on dry surfaces, while Gram-negative bacteria like P. aeruginosa can survive in moist environments such as sinks for extended periods. Moreover, the infective dose of these bacteria appears to be very low, meaning that even slight environmental contamination is sufficient to cause infection. 13. Even though modern techniques for instrument sterilization, improved operating rooms, and great efforts of infection prevention strategies used, still SSI remains as HAI 14.
The burden of SSIs is very high in developing countries, where limited resources, poor infection control practices, overcrowded hospital settings, and inappropriate antimicrobial use are common challenges. Studies conducted in Ethiopia on postoperative SSI have showed the incidence of 9.8%, and 21% in Addis Ababa 15, and Mekelle 16 respectively.
Previously, the operating rooms at the University of Gondar Comprehensive Specialized Hospital were renovated, well-organized, and equipped. However, there is no evidence indicating whether there has been a decrease in surgical site infections or multidrug-resistant bacteria as a result.
Therefore, continuous surveillance is necessary to guide appropriate therapy for surgical site infections and ensure the rational use of antimicrobial agents. This approach is crucial for preventing the emergence of multidrug-resistant pathogens. A recent study is needed to update the current knowledge of etiologic agents and their antimicrobial susceptibility patterns of isolates. Such a study will support clinicians in selecting appropriate treatments and provide insight into the definitive diagnosis of surgical site infections based on local bacterial susceptibility profiles. Additionally, this information is vital for infection prevention and control efforts.
A hospital-based cross-sectional study was conducted at the University of Gondar Comprehensive Specialized Hospital (UoGCSH) from February 1 to April 30, 2020. UoGCSH is one of the largest tertiary-level referral and teaching institutions in the Amhara region, located in Gondar town, approximately 750 km northwest of Addis Ababa, Ethiopia. According to the Central Statistical Agency of Ethiopia report in 2015, Gondar town comprises twelve sub-cities, twenty-two urban and eleven rural kebeles, with a total projected population of 323,900. UoGCSH, as reported by its admission and discharge office, has 700 beds and includes departments for surgical, medical, pediatric, gynecologic, obstetrics, fistula care, and intensive care units. The hospital serves residents of Gondar town, surrounding zones, and neighboring regions.
The study population included all patients who had developed postoperative surgical site infections in the surgical, orthopedics, and gynecology & obstetric wards at UoGCSH during the study period. We collected environmental samples along with wound swabs to assess the similarity between the etiologies of wound isolates and environmental samples isolates. Our hypothesis was that the hospital environment serves as a source of surgical site infections. Therefore, environmental samples were collected from inanimate objects such as bed rails, tray tables, IV poles, bedside tables, room sinks, room light switches, door knobs, as well as air bacteriological samples.
All postoperative patients suspected of having surgical site infections and hospital environments during the study period were included in the study.
All postoperative patients suspected of having surgical site infections were included in the study.
Patients who were very critical and difficult to take samples were excluded.
Socio-demographic characteristics were collected from each study participant through face-to-face interviews using a structured questionnaire. Wound swabs were collected aseptically using sterile cotton-wool swabs soaked in normal saline during dressing changes from the infected surgical site, prior to cleaning with an antiseptic solution. The swabs were then placed into sterile test tubes and immediately transported to the Bacteriology Laboratory 17.
Environmental samples were collected from frequently touched surfaces and equipment in the wards. A sterile cotton swab moistened with sterile normal saline was used for sampling high-touch surfaces. Swabs were taken in the morning, prior to the commencement of routine activities. Each site was swabbed in a close zigzag pattern covering an area of approximately 10 cm², with the swab rotated during sampling to ensure thorough surface coverage 18,19. The swabs were securely placed in labeled sterile tubes and promptly transported to the Bacteriology Laboratory for further processing.
Indoor air samples were collected from the operating rooms and surgical ward units using a settling plate or passive air sampling method. In each operating room, sampling was conducted in the early morning before the start of surgical activities and during surgical procedures on the day. For the wards, air samples were collected in the morning (during healthcare worker rounds) and in the evening (when visitors are present). As per standard procedure, a sterile Petri dish with a diameter of nine centimeters containing 5% Sheep’s blood agar was left open to the air for one hour. This dish was positioned one meter above the floor and one meter from walls or any other obstacles during sampling 20-24.
The swabs collected from the infected surgical sites, surfaces, and equipment were processed immediately upon arrival at the laboratory following standard procedures. Swab samples were inoculated onto MacConkey agar, Mannitol salt agar, Blood agar plates, and Chocolate agar by gently rolling the swab over the agar surfaces. The inoculated agar plates were then incubated at 37°C for 24 to 48 hours. Air samples collected on Blood agar plates for air culture were similarly incubated at 37°C for 24 hours 17.
Presumptive identification of bacteria was done based on its Gram reaction and colony characteristics of the organisms. Confirmatory test was done by enzymatic and biochemical properties of the organisms. Gram-negative rods were identified by performing a series of biochemical tests which include triple sugar iron agar, citrate utilization test, lysine decarboxylase test, indole test, motility test, urease production, and oxidase test while Gram-positive cocci were identified based on their Gram reaction, catalase, coagulase, and bile esculin hydrolyze test 25.
The suspension was prepared from pure isolates using 0.85% normal saline, adjusted to a 0.5 McFarland standard for antimicrobial susceptibility testing. The suspension was evenly distributed on Muller Hinton agar using a sterile cotton applicator stick.
The antimicrobial susceptibility test was conducted using the Kirby-Bauer disk diffusion method, as recommended by the Clinical and Laboratory Standards Institute (CLSI). The following antimicrobials were tested: Cefoxitin (30 μg), Vancomycin (30 μg), Clindamycin (2 μg), Erythromycin (15 μg), Doxycycline (30 μg), Tetracycline (30 μg), Ampicillin (10 μg), Chloramphenicol (30 μg), Gentamycin (10 μg), Ciprofloxacin (5 μg) ; and Trimethoprim-sulphamethoxazole (1.25 / 23.75 μg ) for Gram-positive bacteria.
Similarly, antimicrobial susceptibility test was performed for Gram-negative bacteria using the Kirby-Bauer disk diffusion method for the following antimicrobials: Amikacin (30 μg), Ceftazidime (30 μg), Cefotaxime (30 μg), Cefepime (30 μg), Tobramycin (10 μg), Piperacillin (100 μg) and Meropenem (10 μg), Doxycycline (30 μg), Tetracycline (30 μg), Ampicillin (10 μg), Chloramphenicol (30 μg), Gentamycin (10 μg), Ciprofloxacin (5 μg); and Trimethoprim-sulphamethoxazole (1.25 / 23.75 μg). After applying antimicrobials on Mueller Hinton agar, the plates were incubated for 16-18 hours at 37 °C.The diameter of the zones of inhibition was measured using a ruler. Finally, the results were interpreted as Susceptible, Intermediate, and Resistant using CLSI 2019 26.
The susceptibility of consecutive isolates of S. aureus to Cefoxitin was determined using Muller Hinton agar. Suspension of the overnight growth S. aureus isolate (0.5 McFarland turbidity) was evenly distributed onto Muller Hinton agar. The Cefoxitin (30 μg) disk was aseptically placed on the surface of the inoculated plate and incubated aerobically at 35°C for 16-18 hours. The diameter of the zone of inhibition was measured and compared with CLIS (2019). Cefoxitin (≤ 22 mm diameter) resistant isolates were termed as MRSA 26.
The reliability of the study findings was ensured by implementing stringent quality control measures throughout the entire laboratory process. All materials, equipment, and procedures were thoroughly regulated. Quality assurance was maintained during the pre-analytical, analytical, and post-analytical stages. Additionally, all clinical and environmental specimens were collected in accordance with standard operating procedures. All media were prepared according to the manufacturer’s instructions and standard operating procedures. The sterility of each batch of test medium was confirmed by visually inspecting for growth or discoloration after incubating 5% of uninoculated plates and tubes at 37°C for 24 hours. Media performance was verified by inoculating known control strains. The growth and hemolysis performance on blood agar plates were checked using Staphylococcus aureus ATCC 25923 (β-hemolysis).
MacConkey agar was checked by Staphylococcus aureus ATCC 25923 (no growth) and Escherichia coli ATCC 35218 (lactose fermenter). Mannitol salt agar was checked by Staphylococcus aureus ATCC 25923 (Mannitol fermenter). Additionally, all the aforementioned reference strains were used to checked the quality of the antimicrobial disks26.
Socioeconomic and data obtained from laboratory results were entered and analyzed using the Statistical Package for Social Sciences (SPSS) version 20. Descriptive statistics were calculated to summarize demographic and clinical characteristics. Frequency distribution was used to compute the results. Study findings were presented in words, numbers, percentages, tables and graphs.
A total of 52 study participants who had developed postoperative surgical site infections were included in this study. Of the wound swabs collected, 44 (87.3%) were positive for bacteria, while 8 (12.7%) showed no bacterial growth. Twenty-nine (55.8%) participants were male. The mean age of the participants was 33.8 years with a standard deviation of 13.5 years, ranging from 17 to 75 years. Most participants (46.2%) were in the 21 to 30-year age group. The majority (63.5%) stayed in the hospital for 10-20 days. The abdomen was the most common surgical site (65.4%), and emergency surgeries were the most frequent type of case (67.3%). (Table 1).
Table 1 Socio-demographic and clinical characteristics of the study participants in UoGCSH, Northwest Ethiopia, February – April 2020 (n=52)
| Characteristics | Surgical units | Total No (%) | |||
|---|---|---|---|---|---|
| Surgical ward No (%) | Orthopedic ward No (%) | Gynecology & obstetrics Ward No (%) | |||
| Sex | Male | 15(28.8%) | 14(26.9%) | 0(0.0%) | 29(55.8%) |
| Female | 8(15.4%) | 0(0.0%) | 15(28.8%) | 23(44.2%) | |
| Age in years | 20-Nov | 4(7.7%) | 1(1.9%) | 1(1.9%) | 6(11.5%) |
| 21-30 | 8(15.4%) | 6(11.5%) | 10(19.2%) | 24(46.2%) | |
| 31-40 | 6(11.5%) | 3(5.8%) | 2(3.8%) | 11(21.2%) | |
| >41 | 5(9.6%) | 4(7.7%) | 2(3.8%) | 11(21.2%) | |
| Residence | Rural | 18(34.6%) | 10(19.2%) | 7(13.5%) | 35(67.3%) |
| Urban | 5(9.6%) | 4(7.7%) | 8(15.4%) | 17(32.7%) | |
| Hospital stays (in days) | <10 | 10(19.2%) | 4(7.7%) | 3(5.8%) | 17(32.7%) |
| 20-Oct | 13(25.0%) | 8(15.4%) | 12(23.1%) | 33(63.5%) | |
| 21-30 | 0(0.0%) | 2(3.8%) | 0(0.0%) | 2(3.8%) | |
| Site of operation | Back | 1(1.9%) | 0(0.0%) | 0(0.0%) | 1(1.9%) |
| Thorax | 1(1.9%) | 0(0.0%) | 0(0.0%) | 1(1.9%) | |
| Abdomen | 16(30.8%) | 4(7.7%) | 14(26.9%) | 34(65.4%) | |
| Leg | 2(3.8%) | 7(13.5%) | 0(0.0%) | 9(17.3%) | |
| Other* | 3(5.8%) | 3(5.8%) | 1(1.9%) | 7(13.5%) | |
| Type of cases | Emergency | 12(23.2%) | 11(21.2%) | 12(23.1%) | 35(67.3%) |
| Elective | 11(21.2%) | 3(5.8%) | 3(5.8%) | 17(32.7%) | |
Approximately 44 (84.6%) clinical samples were culture-positive. Among these, 33 (75%) had single isolates, while 11 (25%) had mixed isolates. The highest number of isolates was found in patients admitted to the surgical ward (24, 43.6%), followed by those in the orthopedics ward (16, 29.1%). Additionally, a total of 150 environmental samples (104 surface and 46 air samples) were collected from the wards, with 116 (77.3%) testing positive for bacterial culture. The predominant isolates were CoNS 57(47.5%) followed by S. aureus 35(29.2%), Klebsiella spp 7(5.8%) and P. aeroginosa and E.coli each 6(8.1%). Of those 46 air samples, 44(95.7%) were culture-positive. Similarly, CoNs were the most common isolates in air samples 22(47.8%), followed by S. aureus 17(37.0%), Enterococcus spp 3(6.5%), and E. cloacae 1(2.2%) (Table 2).
Table 2 Distribution of isolated bacteria in different surgical units and hospital environments at UoGCSH, Northwest, Ethiopia, February – April 2020
| Bacterial Isolates | Surgical units | Total N (%) | Hospital environments | Total N (%) | Over All Total N (%) | |||
|---|---|---|---|---|---|---|---|---|
| Surgical ward N (%) | Orthopedic ward N (%) | Gynecology & obstetrics ward N (%) | Surfaces samples N (%) | Air samples N (%) | ||||
| S. aureus | 4 (7.3) | 1(1.9) | 6(10.9) | 11 (20.0) | 18 (24.3) | 17 (37.0) | 35 (29.2) | 46 (26.3) |
| CoNS | 1(1.9) | 2 (3.6) | 2(3.6) | 5 (9.1) | 35 (47.3) | 22 (47.8) | 57 (47.5) | 62 (35.4) |
| Enterococcus spp | 2 (3.6) | 4 (7.3) | 0 (0.0) | 6 (10.9) | 2(2.7) | 3(6.5) | 5(4.2) | 11 (6.3) |
| Klebsiella spp | 5 (9.1) | 3 (5.5) | 3 (5.5) | 11 (20.0) | 4 (5.4) | 3 (6.5) | 7 (5.8) | 18 (10.3) |
| E. coli | 6 (10.9) | 4 (7.3) | 0 (0.0) | 10 (18.2) | 6 (8.1) | 0 (0.0) | 6 (5.0) | 16 (9.1) |
| E. cloacae | 1 (1.9) | 1 (1.9) | 3 (5.5) | 5 (9.1) | 2 (2.7) | 1(2.2) | 3 (2.5) | 8 (4.6) |
| P. aeroginosa | 5 (9.15) | 0 (0.0) | 0 (0.0) | 5 (9.1) | 6(8.1) | 0(0.0) | 6 (5.0) | 11 (6.3) |
| Acinitobacter spp | 0 (0.0) | 1 (1.9) | 1 (1.9) | 2 (3.6) | 1 (1.4) | 0 (0.0) | 1 (0.8) | 3 (1.7) |
| Total | 24 (43.6) | 16 (29.1) | 15 (27.3) | 55 (100) | 74 (100) | 46 (100) | 120 (100) | 175 (100) |
Thirty-three (60%) of the isolates were Gram-negative bacteria, while 22 (40%) were Gram positive. The most common isolates were S. aureus and Klebsiella spp., each accounting for 11 (20%) of the total isolates, followed by E. coli with 10 (18.2%) isolates (Figure 1).

The distribution of bacterial isolates varied among the wards. Most isolates were obtained from surgical wards (38, 31.7%). Staphylococcus aureus (17, 14.2%) and E. coli (3, 2.5%%) were most frequently found in surgical wards compared to other wards respectively. Enterococcus species (4, 3.3%) and Klebsiella species (4, 3.3%) also isolated most frequently from surgical wards. Coagulase-negative staphylococci isolates were most frequently isolated from surgical ward (23, 19.1%) and orthopedic wards (19,15.8%) followed by gynecology and obstetrics ward (12.5%) (Table 3).
Table 3 Distribution of isolated bacteria from environmental samples among wards in UoGCSH, Northwest Ethiopia, February – April 2020
| Isolated bacteria | Surgical units | Total N (%) | ||
|---|---|---|---|---|
| Surgical ward | Orthopedic ward | Gynecology & obstetrics ward | ||
| N (%) | N (%) | N (%) | ||
| S. aureus | 17(14.2%) | 10(8.3%) | 8(6.7%) | 35(29.2%) |
| Klebsiella spp | 4(3.3%) | 1(0.8%) | 2(1.7%) | 7(5.8%) |
| E. coli | 3(2.5%) | 2(1.7%) | 1(0.8%) | 6(5.0%) |
| Pseudomonas spp | 4(3.4%) | 1(0.8%) | 1(0.8%) | 6(5.0%) |
| E. cloacea | 1(0.8%) | 1(0.8%) | 1(0.8%) | 3(2.5%) |
| Acinetobacter spp | 1(0.8%) | 0(0.0%) | 0(0.0%) | 1(0.8%) |
| Cons | 23(19.1%) | 19(15.8%) | 15(12.5%) | 57(47.5%) |
| Enterococcus spp | 4(3.3%) | 1(0.8%) | 0(0.0%) | 5(4.1%) |
| Total No. (%) | 57(47.5%) | 35(29.2%) | 28(23.3%) | 120(100%) |
The antimicrobial susceptibility patterns of Gram-positive bacteria isolated from surgical sites and hospital environments are presented in Table 4. Staphylococcus aureus from surgical sites showed high resistance rates to Penicillin (90.9%), Tetracycline (90%), Erythromycin (72.7%), and Cotrimoxazole (72.7%). The MRSA isolation rate in this study was 63.6%, as indicated by Cefoxitin resistance. All six Enterococcus isolates were susceptible to Vancomycin.
Regarding environmental isolates S. aureus exhibited resistance to Cotrimoxazole (60%) and Penicillin (80%), while showing sensitivity to Clindamycin (100%) and Ciprofloxacin (91.4%). The prevalence of MRSA was 17(48.6). All of the five Enterococcus spps were Vancomycin susceptible (Table 4).
Table 4 Antimicrobial-resistant pattern of Gram-positive bacterial isolates from surgical sites and Hospital environments in UoGCSH, Northwest Ethiopia, February – April 2020
| Antimicrobial agent | Gram-positive bacteria | |||||
|---|---|---|---|---|---|---|
| S. aureus (N, %) | CoNS (N, %) | Enterococcus spp (N, %) | ||||
| Patient (N, %) | Hospital environment (N, %) | Patient (N, %) | Hospital environment (N, %) | Patient (N, %) | Hospital environment (N, %) | |
| Ciprofloxacin | 4(36.4%) | 3(8.6%) | 3(60%) | 9(15.8%) | _ | _ |
| Chloramphenicol | _ | 12(34.3%) | _ | 10(17.5%) | 3(50%) | 2(40.0%) |
| Gentamycin | 6(54.5%) | 12(34.3%) | 1(20%) | 18(31.6%) | _ | _ |
| Cotrimoxazole | 8(72.7%) | 21(60%) | 4(80%) | 35(61.4%) | _ | _ |
| Tetracycline | 10(90.9%) | _ | 3(60%) | _ | _ | _ |
| Erythromycin | 8(72.7%) | 12(34.3%) | 3(60%) | 25(43.9%) | _ | _ |
| Clindamycin | 2(18.2%) | 0(0.0%) | 1(20%) | 8(14.0%) | _ | _ |
| Penicillin | 10(90.9%) | 28(80%) | 5(100%) | 49(86.0%) | 6(100%) | 4(80.0%) |
| Ampicillin | _ | _ | _ | _ | 5(83.3%) | 1(20.0%) |
| Cefoxitin | 7(63.3%) | 17(48.6%) | 3(60%) | 25(43.9%) | _ | _ |
| Vancomycin | _ | _ | _ | _ | 0(0.0%) | 0(0.0%) |
| Doxycycline | 9(81.8%) | 13(37.1%) | 2(40%) | 14(24.6%) | 6(100%) | 3(60.0%) |
Table 5 presents the antimicrobial susceptibility patterns of Gram-negative bacteria isolated from surgical sites and hospital environments. Klebsiella spp. exhibited resistance rates of 100% to Cefotaxime and 81.8% to both Ceftazidime and Cotrimoxazole. E. coli showed 100% resistance to Cefotaxime and Cotrimoxazole, and 80% resistance to Ceftazidime. Pseudomonas isolates demonstrated resistance rates of 80% to Ceftazidime, and 60% to Ciprofloxacin, Gentamycin, and Tobramycin. In contrast, among environmental isolates, Klebsiella spp. showed high resistance rates of 100% to both Cotrimoxazole and Cefotaxime. Similarly, E. coli exhibited 100% resistance to both Cotrimoxazole and Cefotaxime. Pseudomonas isolates from the environment were resistant to Ciprofloxacin and Tobramycin, each at 83.3%.
Table 5 Antimicrobial resistant pattern of Gram-negative bacterial isolates from surgical sites and Hospital environments in UoGCSH, Northwest Ethiopia, February – April 2020
| Antimicrobial agent | Gram-negative bacteria | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Klebsiella spp N (%) | E. coli N (%) | P. aeroginosa N (%) | E. cloacae N (%) | Acinitobacter spp N (%) | ||||||
| Patient N(%) | Evnt. N( %) | Patient N( %) | Envt. N( %) | Patient N( %) | Envt. N( %) | Patient N( %) | Envt. N(%) | Patient N(%) | Envt. N (%) | |
| CIP | 3(27.3) | 2(28.6) | 4(40) | 4(66.7) | 3(60) | 5(83.3) | 2(40) | 0(0.0) | 0(0.0) | _ |
| CAF | 3(27.3) | 5(71.4) | 2(20) | 5(83.3) | _ | _ | 3(60) | 1(33) | _ | _ |
| GEN | 4(36.4) | 5(71.4) | 2(20) | 4(66.7) | 3(60) | 3(50) | 3(60) | 2(66.7) | 1(50) | 1(100) |
| COT | 9(81.8) | 7(100) | 10(100) | 6(100) | _ | _ | 3(60) | 1(33.3) | 1(50) | 1(100) |
| CAZ | 9(81.8) | 6(85.7) | 8(80) | 3(50) | 4(80) | 3(50) | 2(40) | 0(0.0) | 1(50) | 1(100) |
| CXT | 11(100) | 7(100) | 10(100) | 6(100) | _ | _ | 4(80) | 3(100) | 2(100) | 1(100) |
| TOB | 7(63.6) | 4(57.1) | 3(30) | 6(100) | 3(60) | 5(83.3) | 4(80) | 1(33.3) | 1(50) | 0(0.0) |
| AMK | 1(9.1) | 0(0.0) | 1(10) | 0(0.0) | 1(20) | 0(0.0) | 1(10) | 0(0.0) | 0(0.0) | 0(0.0) |
| MER | 7(63.6) | 2(28.6) | 2(20) | 0(0.0) | 2(40) | 2(33.3) | 3(60) | 0(0.0) | 1(50) | _ |
| PEP | _ | _ | _ | _ | 2(40) | 3(50) | _ | _ | _ | |
| CEF | _ | _ | _ | _ | 2(40) | 4(66.7) | _ | _ | _ | |
The overall MDR resistance from surgical sites was observed in 31 cases (56.4%). Among these, 15 (68.2%) were from Gram-positive isolates and 16 (48.5%) from Gram-negative isolates. Of the 11 S. aureus isolates, 9 (81.8%) were found to be MDR. (Table 6).
Table 6 The multidrug-resistant pattern of bacterial isolates from surgical sites in UoGCSH, Northwest Ethiopia, February – April 2020
| Bacterial isolate | Total | Multidrug-resistant pattern | ||||||
|---|---|---|---|---|---|---|---|---|
| R0 | R1 | R2 | R3 | R4 | ≥R5 | Total MDR | ||
| Gram-positive bacteria | 22 | 0 | 2 | 8 | 4 | 2 | 9 | 15(68.2%) |
| S. aureus | 11 | 0 | 1 | 1 | 0 | 2 | 7 | 9(81.8%) |
| Cons | 5 | 0 | 0 | 2 | 1 | 0 | 2 | 3(60%) |
| Enterococcus spp | 6 | 0 | 0 | 3 | 3 | 0 | 0 | 3(50%) |
| Gram-negative bacteria | 33 | 3 | 5 | 11 | 9 | 4 | 3 | 16(48.5%) |
| Klebsiella spp | 11 | 0 | 2 | 4 | 1 | 2 | 2 | 5(45.5%) |
| E. coli | 10 | 0 | 1 | 3 | 5 | 1 | 0 | 6(60%) |
| E. cloacae | 5 | 1 | 0 | 1 | 1 | 1 | 1 | 3(60%) |
| Pseudomonas spp | 5 | 0 | 2 | 1 | 2 | 0 | 0 | 2(40%) |
| Acinetobacter spp | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Total | 55 | 3 | 7 | 19 | 13 | 6 | 12 | 31(56.4%) |
The overall multidrug-resistant (MDR) resistance from hospital environments was 66 cases (55.0%). Among these, 53 (54.6%) were from Gram-positive isolates, and 13 (56.5%) were from Gram-negative isolates. More than 70% of Klebsiella isolates were multidrug-resistant (see Table 7).
Table 7 The multidrug-resistant pattern of bacterial isolates from environments at the UoGCSH, Northwest Ethiopia, February – April 2020
| Bacterial isolates | Total | Multidrug-resistant pattern | ||||||
|---|---|---|---|---|---|---|---|---|
| R0 | R1 | R2 | R3 | R4 | ≥R5 | Total MDR | ||
| Gram-positive bacteria | 97 | 9 | 27 | 8 | 11 | 10 | 32 | 53(54.6%) |
| S. aureus | 35 | 2 | 11 | 3 | 4 | 3 | 12 | 19(54.3%) |
| CoNS | 57 | 6 | 13 | 4 | 7 | 7 | 20 | 34(59.6%) |
| Enterococcus spp | 5 | 1 | 3 | 1 | 0 | 0 | 0 | 0 |
| Gram-negative bacteria | 23 | 2 | 1 | 7 | 8 | 1 | 4 | 13(56.5%) |
| Klebsiella spp | 7 | 0 | 0 | 2 | 3 | 0 | 2 | 5(71.4%) |
| E. coli | 6 | 0 | 0 | 2 | 3 | 1 | 0 | 4(66.7%) |
| Pseudomonas spp | 6 | 0 | 1 | 2 | 1 | 0 | 2 | 3(50.0%) |
| E. cloacae | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| Acinetobacter spp | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1(100%) |
| Total | 120 | 11 | 28 | 15 | 19 | 11 | 36 | 66(55.0%) |
The present study provides insights into the bacterial profiles and their antimicrobial susceptibility patterns in samples from postoperative surgical site infections and hospital environments, crucial for selecting appropriate antimicrobial agents and preventing future infections. The culture positivity rate for postoperative SSIs was 84.6%, with the main isolates being S. aureus (20%), Klebsiella spp. (20%), E. coli (18.2%), Enterococcus spp. (10.9%), and Pseudomonas spp. (9.1%). These findings are consistent with a study conducted six years ago at the same hospital in Gondar, Ethiopia, where S. aureus (22.4%) and Klebsiella spp. (20.4%) were the predominant isolates 27. This result is also consistent with studies conducted in Hawassa, Ethiopia 28, Mekelle, Ethiopia 29, Pakistan 8,30, and Saudi Arabia 31 where S. aureus, Klebsiella spp, and E. coli were reported as major isolates of SSIs. However, this contrasts with findings from studies in Tanzania and India, where Pseudomonas species were found to be the predominant isolates 32,33.
In this study, Klebsiella species and E. coli also showed high prevalence, similar to S. aureus which could be due to contamination of the wound with the gastrointestinal tract in which they are normal floras and most operations were undertaken on the abdomen.
In the present study, Gram-positive bacteria were found in 40% and Gram-negative bacteria in 60% of culture-positive cases of SSIs. Similar studies conducted in different regions have also highlighted that Gram-negative bacteria are frequently identified as a more significant cause of SSIs compared to Gram-positive bacteria 31,34,35. This could be attributed to the diverse habitats of Gram-negative bacteria, including inanimate surfaces in hospitals, and potential contamination from the intestinal tract during surgery. S. aureus typically plays a predominant role in HAI due to contamination of wounds with normal endogenous flora found on the skin and mucous membranes, or through environmental contamination. S. aureus can survive for extended periods on dry surfaces in hospital environments 36. Due to its ability to survive for extended periods in hospital environments, it contributes to the emergence of drug-resistant strains.
The prevalence of MRSA in this study (63.6%) was consistent with a study conducted in Pakistan (65.7%) 8. On the contrary, this finding was higher than the results of studies conducted in Gondar, Ethiopia (34.7%) 34, Debre Markos, Ethiopia (49.7%) 37, Addis Ababa, Ethiopia (10.5%) 15, Nepal (41.7%) 38 and India (48.8%) 39. The high incidence rate of MRSA in this study could be attributed to improper antimicrobial use or the prophylactic use of antimicrobials, leading to the timely emergence of resistant strains. Most of the isolates of S. aureus (81.8%) were Clindamycin susceptible which is in agreement with the study conducted in Gondar 34 and Saudi Arabia 31 in which 88.5% and 97.2% of the isolates were clindamycin susceptible correspondingly.
Our results have shown that Amikacin was effective against more than 80% of Gram-negative bacterial isolates. However, these isolates exhibited high resistance rates to Cefotaxime, Cotrimoxazole, and Ceftazidime (ranging from 72.7% to 82.1%). Klebsiella species demonstrated high resistance rates to Cefotaxime (100%), Cotrimoxazole, and Ceftazidime (81.8% each). Similarly, E. coli was resistant to Cefotaxime and Cotrimoxazole (100% each) and Ceftazidine (80%). These findings are in agreement with the study done in Addis Ababa, Ethiopia 15 which showed that Klebsiella spp and E. coli were highly resistant to Ceftazidime (80%, 79.2). Another study done in Addis Ababa, Ethiopia 40 also showed Ceftazidime and Cotrimoxazole were not effective antimicrobials for E. coli and Klebsiella spp. Similarly, Pseudomonas isolates showed resistance to Ceftazidime (80%), Ciprofloxacin, Gentamycin, and Tobramycin (60% each). It was observed that all surgery patients in the study area received Ceftriaxone as prophylaxis, which likely contributed to the emergence of resistant bacteria.
In the current study, 56.4% of the isolates were MDR. This result was lower than the result of the studies conducted in Addis Ababa 35, in which the MDR level was 65.5%, and Nepal 38 also showed 66.7% of MDR. This variation might be due to the difference in the definition of MDR between the two studies. In previous studies, MDR was defined as resistance to two or more classes of antimicrobials and improper antimicrobial usage practices in the respective areas.
Several studies have demonstrated that healthcare facility environments, including frequently touched surfaces and air, are contaminated by various types of bacteria, contributing to HAI 41-43. In the current study, the overall contamination rate of the hospital environment was 116 (77.3%), with contamination rates of 69.2% among 44 inanimate surfaces and 95.7% among air samples. This finding aligns with studies conducted in Nepal and Brazil 45 which reported contamination rates of 78% and 83.3%, respectively. However, it was higher than studies conducted in Mizan Tepi, Ethiopia43 (43.8%), Uganda 42 (44.2%), and Poland 19 (69.6%) (19). The variation may be due to differences in cleaning practices, decontamination of surfaces, and the effectiveness of disinfectant use.
In this study, hospital environment isolates of S. aureus showed 100% susceptibility to Clindamycin and 91.4% susceptibility to Ciprofloxacin. Of all S. aureus isolates, 17 (48.6%) were MRSA, which is higher than the rate reported in a study conducted in Bahir Dar (25%) 41. The variation may be attributed to differences in infection prevention practices between the two settings or variations in antimicrobial use for treating bacterial infections across different hospitals. The overall MDR level of the isolates in this study was 66 (55.0%), which is lower than the study conducted in Bahir Dar 41, where more than 75% of isolates were reported to be MDR.
Furthermore, among the total Gram-positive bacterial environmental isolates, 53 (54.6%) were multidrug-resistant (MDR) in our study. Additionally, 13 (56.5%) of the gram-negative isolates were also MDR. Klebsiella spp. and E. coli isolates in our study showed resistance to Cotrimoxazole and Cefotaxime but were susceptible to Amikacin which the primary causative bacteria associated with postoperative surgical site infections (SSI), and hospital environments served as potential reservoirs for these pathogens in the study area.
Staphylococcus aureus, Klebsiella species, and E. coli were identified as the most prevalent bacteria from postoperative surgical site infections, with hospital environments serving as potential reservoirs for these pathogens in the study area. The study also revealed a high prevalence of methicillin-resistant and multidrug-resistant strains among clinical and hospital isolates. However, Amikacin and Clindamycin demonstrated effectiveness in inhibiting the in vitro growth of Gram-negative and Gram-positive bacterial isolates, respectively. To curb the further emergence and spread of multidrug-resistant bacterial pathogens, treatment guidelines for the use of antimicrobials should be updated based on the hospital formularies and the susceptibility patterns. Additionally, infection prevention practices should be strengthened.
We express our heartfelt gratitude to the study participants and the staff of the University of Gondar Comprehensive Specialized Referral Hospital, particularly those working at the tuberculosis laboratory and bacteriology teaching laboratory. Their significant contributions throughout the data and specimen collection process are deeply appreciated.
CLSI: Clinical and Laboratory Standards Institute, CoNS: Coagulase-negative Staphylococci, HAIs: Health-care-associated infections, MDR: Multidrug-resistant, MRSA: Methicillin-resistant Staphylococcus aureus, SSI Surgical site infection, UoGCSH: University of Gondar Comprehensive Specialized Hospital, WHO: World Health Organization
This study received approval from the Ethical Review Committee of the School of Biomedical and Laboratory Sciences, University of Gondar. Informed consent was obtained from each study participant and their legal guardians after explaining the study's purpose. Participant information was treated confidentially, and specimens collected were used solely for the study's intended purposes. All procedures in this study were conducted in accordance with the amended Declaration of Helsinki.
Not applicable.
All the data sets analyzed during the current study are available from the corresponding author upon reasonable request.
The authors declare that they have no competing interests.
There is no specific fund received for this study.
SB and AA contributed to conceiving the research idea, data collection, and data analysis. GB and WT contributed to the conception of the research idea, method rationalization, data analysis, interpretation of results, evaluation of scientific content, and manuscript preparation. GB, SB, FW, and WA were also involved in reviewing and editing the manuscript. All authors have read and approved the final manuscript for submission.